
 |
||||||||

| Tracks 1-19 | ||||||||||||||||||||||||||||||||||||||||
|
Select Excerpts from the Roundtable
Tracks 2-4
![]() DR LOVE: Lisa, can you review the landmark work by your colleague Charles Perou and others on the molecular classification
of breast cancer?
DR LOVE: Lisa, can you review the landmark work by your colleague Charles Perou and others on the molecular classification
of breast cancer?
![]() DR CAREY: It turns out that molecular portraits of breast cancer differ from one type to another. At least five subtypes, and probably more, exist.
DR CAREY: It turns out that molecular portraits of breast cancer differ from one type to another. At least five subtypes, and probably more, exist.
The seminal paper on this topic was coauthored by Chuck Perou and Therese Sørlie. It was called “Molecular Portraits of Human Breast Tumours.” They asked simple questions: Can you identify a subset of genes in a gene expression array that seem to be more altered in one group than another group of breast cancers? Is breast cancer a range of different diseases, or are clusters identifiable (Perou 2000)?
They performed cluster analysis to identify whether groups of breast cancer existed. They started with a relatively small dataset of fewer than 100 samples (Perou 2000). They asked: Which genes are the most altered between cancers but not altered within cancers?
Within that dataset, a group of breast cancer samples were collected before and after neoadjuvant chemotherapy. They said, “We want to eliminate genes that may have changed in the same woman, before and after doxorubicin, and keep the genes that are the most different between cancers” (Perou 2000).
They ended up with about 500 genes that met those criteria. About a fourfold difference was identified between one cancer and another, and they tended to stay the same within a cancer. That’s where the 496 genes — what’s called the intrinsic list — came from (Perou 2000).
The five subtypes we talk about most commonly include the two luminal subtypes — luminal A and luminal B, the basal-like subtype (Perou 2000; Sørlie 2001), the HER2 subtype, which is what we now call the HER2-positive/ER-negative subtype, and then the unclassifiable group that’s called the normal-like type, probably because there was too much stroma involved to categorize them effectively (Perou 2000; Sørlie 2001).
These profiles have been replicated in multiple independent data sets. They exist regardless of treatment and outcome, which is not to say they don’t have prognostic implications. They do, and that’s also been shown in multiple independent datasets.
If there were no prognostic relevance of these subtypes, it wouldn’t change their value. I don’t believe they’re the best way to prognosticate, and they weren’t developed for prognostication. Their value is in their ability to biologically define the different diseases under the umbrella of breast cancer.
![]() DR CHANG: I believe that’s absolutely true — the molecular classification just tells you the different subtypes of breast cancer. For me, what would be interesting to determine is whether all these subtypes come from the same cell of origin.
DR CHANG: I believe that’s absolutely true — the molecular classification just tells you the different subtypes of breast cancer. For me, what would be interesting to determine is whether all these subtypes come from the same cell of origin.
In terms of what we know, there are five broad categories, and maybe more, perhaps, in the triple-negative groups. There may be more than the basal type, and there may be different types within that.
Track 5
![]() DR LOVE: Can you review the subtypes in terms of HER2, ER and other biological and clinical characteristics?
DR LOVE: Can you review the subtypes in terms of HER2, ER and other biological and clinical characteristics?
![]() DR CAREY: Luminal A and luminal B are the two ER-positive subtypes. The luminal As tend to have a higher expression of the ER and ER-regulated genes compared to the luminal Bs, which tend to have a lower expression of ER and related genes (Sørlie 2001).
DR CAREY: Luminal A and luminal B are the two ER-positive subtypes. The luminal As tend to have a higher expression of the ER and ER-regulated genes compared to the luminal Bs, which tend to have a lower expression of ER and related genes (Sørlie 2001).
The HER2-positive, ER-positive subtypes tend to fall into the luminal B category, which tends to have a higher expression of the proliferative gene clusters.
![]() DR LOVE: Why are they called “luminal”?
DR LOVE: Why are they called “luminal”?
![]() DR CAREY: The gene expression pattern they most resemble from the normal epithelial component is the luminal epithelial cells. The basal-like subtypes, similarly, have a crude resemblance to the expression patterns of the basal epithelial cells of the breast (Perou 2000).
DR CAREY: The gene expression pattern they most resemble from the normal epithelial component is the luminal epithelial cells. The basal-like subtypes, similarly, have a crude resemblance to the expression patterns of the basal epithelial cells of the breast (Perou 2000).
The basal-like subtype we call the triple-negatives in the clinical scenario because they’re usually low in ER and its related genes and low in HER2. When we conduct clinical assays, most ER-negative, PR-negative, HER2-negative breast cancers are basal-like. The proliferative gene cluster is usually high in this subtype. The HER2 subtype is high in the genes that are related to HER2 expression and usually low in the ER-related genes (Perou 2000).
![]() DR CHANG: Work that Craig Allred has been doing indicates that the same classification holds true for DCIS. Considering the natural evolution of the disease, DCIS has the same five subgroups, and the fact that DCIS has a similar molecular profile as invasive cancer is extremely interesting.
DR CHANG: Work that Craig Allred has been doing indicates that the same classification holds true for DCIS. Considering the natural evolution of the disease, DCIS has the same five subgroups, and the fact that DCIS has a similar molecular profile as invasive cancer is extremely interesting.
Track 7
![]() DR LOVE: Can you review what we know about the mechanism of action of chemotherapy?
DR LOVE: Can you review what we know about the mechanism of action of chemotherapy?
![]() DR CHANG: Essentially, chemotherapy
affects dividing cells as well as the apoptotic pathway. A cancer cell proliferates, grows and divides because it can beat the apoptotic death signals. Chemotherapy targets cells that divide quickly and, therefore, there will be an increase in apoptosis affecting primarily dividing cells.
DR CHANG: Essentially, chemotherapy
affects dividing cells as well as the apoptotic pathway. A cancer cell proliferates, grows and divides because it can beat the apoptotic death signals. Chemotherapy targets cells that divide quickly and, therefore, there will be an increase in apoptosis affecting primarily dividing cells.
The apoptotic pathway, primarily, is the PI3 kinase AKT pathway. There are several upstream receptors and ligands that feed into this pathway. By and large, chemotherapy has a shotgun approach in affecting this pathway, and the apoptotic death signals will affect most dividing cells. That is why you have nonspecific toxicities associated with chemotherapy, including alopecia, GI toxicity, et cetera.
The trick now is whether we can find specific portraits that would distinguish different tumors so that they can receive specific therapeutic agents.
We have completed a study involving approximately 120 patients who have been randomly assigned to receive either a taxane or an anthracycline. The data are not published, but the portraits are very different. We have a nice, robust signature now for taxanes, as well as for anthracyclines, and they’re different.
Track 10
![]() DR LOVE: Can you provide an update on HER2-positive breast cancer, focusing on the pathways involved, the interaction with the ER pathway and how trastuzumab
and lapatinib affect the cells?
DR LOVE: Can you provide an update on HER2-positive breast cancer, focusing on the pathways involved, the interaction with the ER pathway and how trastuzumab
and lapatinib affect the cells?
![]() DR CAREY: I consider HER2-driven breast cancer, in a biologic sense, as being at least two different groups. The HER2-positive, hormone receptor-negative group is different from the HER2-positive, hormone receptor-positive group. They both benefit from HER2-targeted treatments,
but they are different.
DR CAREY: I consider HER2-driven breast cancer, in a biologic sense, as being at least two different groups. The HER2-positive, hormone receptor-negative group is different from the HER2-positive, hormone receptor-positive group. They both benefit from HER2-targeted treatments,
but they are different.
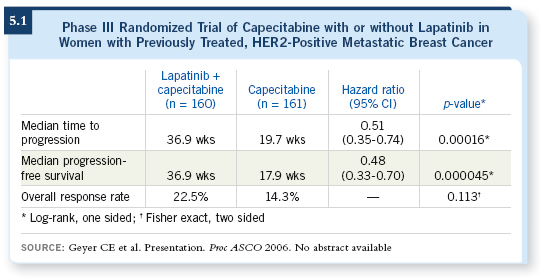
In terms of how HER2 functions, we’re obtaining a lot of information from the emerging studies of trastuzumab resistance and the pathways that are important in trastuzumab resistance. The first issue — and I believe lapatinib speaks to this — is whether HER1 is important in acquired HER2 resistance.
The fact that lapatinib shows efficacy in patients with acquired trastuzumab resistance (Geyer 2006; [5.1]), I believe, provides a strong suggestion that the HER1 pathway may be implicated in getting around HER2 signaling. Tumor cells are smart, and they figure out ways to go around our therapeutic interventions.
![]() DR CHANG: A large component of our work is evaluating the cross talk between the estrogen receptor and HER2. Increasing evidence shows that if you block the HER2 pathway, you can actually upregulate ER, and vice versa. There is crosstalk between the two, and this may be another escape mechanism for trastuzumab resistance.
DR CHANG: A large component of our work is evaluating the cross talk between the estrogen receptor and HER2. Increasing evidence shows that if you block the HER2 pathway, you can actually upregulate ER, and vice versa. There is crosstalk between the two, and this may be another escape mechanism for trastuzumab resistance.
Tracks 11-12
![]() DR LOVE: Jenny, can you discuss cMYC and TOPO II?
DR LOVE: Jenny, can you discuss cMYC and TOPO II?
![]() DR CHANG: Soon Paik presented his data on cMYC. The bottom-line, take-home message was: If you have HER2-positive and cMYC-positive disease, you do well with trastuzumab-based therapies (Kim 2005). This is not the result we expected. cMYC is an oncogene — it basically feeds into the survival pathway. It was expected that if you had cMYC-positive disease, you would do very badly.
DR CHANG: Soon Paik presented his data on cMYC. The bottom-line, take-home message was: If you have HER2-positive and cMYC-positive disease, you do well with trastuzumab-based therapies (Kim 2005). This is not the result we expected. cMYC is an oncogene — it basically feeds into the survival pathway. It was expected that if you had cMYC-positive disease, you would do very badly.
In the adjuvant trastuzumab study, however, patients with cMYC-positive disease who received trastuzumab did extremely well. Their chance of relapsing was very low, less than 10 percent (Kim 2005). This was counterintuitive, probably because trastuzumab works through the PI3 kinase AKT survival pathway and, somehow, cMYC is synergistic because it affects the same pathway. That was an unexpected result of the study.
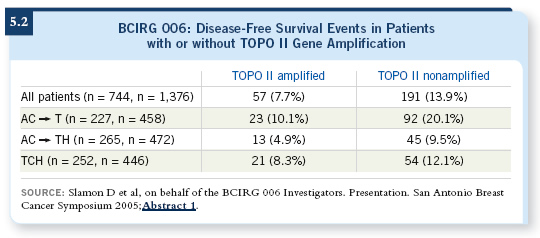
TOPO II is a slightly different story. It is the target for anthracyclines. We know trastuzumab in combination with anthracyclines adversely affects cardiac function and increases cardiotoxicity. Therefore, they wanted to determine whether there were subpopulations of patients receiving trastuzumab who could be spared therapy with anthracyclines.
As presented at the 2005 San Antonio Breast Cancer Symposium, the study demonstrated that, across the board, the nonanthracycline-containing trastuzumab-based regimen was not superior to anthracycline-containing trastuzumab-based therapy. The subset of patients with TOPO II nonamplified disease who received a nonanthracycline-containing regimen, however, did as well as those who received anthracyclines (Slamon 2005; [5.2]).
Whether there is a “smart” population of patients with TOPO II-positive disease who would benefit from anthracyclines and a TOPO II-negative population that could benefit from the absence of anthracyclines is something that needs to be studied.
Track 16
![]() DR LOVE: Lisa, can you review the current Intergroup neoadjuvant
study and how this relates to translational research?
DR LOVE: Lisa, can you review the current Intergroup neoadjuvant
study and how this relates to translational research?
![]() DR CAREY: In the trial for patients with HER2-positive breast cancer, all patients will receive weekly paclitaxel combined with trastuzumab, lapatinib or both in the neoadjuvant setting. The patients all undergo surgery, and in the postoperative period, they receive dose-dense AC and a year of trastuzumab.
DR CAREY: In the trial for patients with HER2-positive breast cancer, all patients will receive weekly paclitaxel combined with trastuzumab, lapatinib or both in the neoadjuvant setting. The patients all undergo surgery, and in the postoperative period, they receive dose-dense AC and a year of trastuzumab.
The primary endpoint is predicated on the idea that the pathologic complete response rate will be higher for the combination than either single agent alone.
![]() DR LOVE: Do you think people will wait for the data from this study before bringing lapatinib into trials
in the adjuvant setting?
DR LOVE: Do you think people will wait for the data from this study before bringing lapatinib into trials
in the adjuvant setting?
![]() DR CAREY: No, I don’t think so. Lapatinib is such an interesting drug that the adjuvant trials will be conducted concurrently with the neoadjuvant trials.
DR CAREY: No, I don’t think so. Lapatinib is such an interesting drug that the adjuvant trials will be conducted concurrently with the neoadjuvant trials.
We will collaborate with the European trial that José Baselga is conducting, which is called Neo-Aphrodite. It has a similar trial design, except their adjuvant chemotherapy regimen is FEC and the adjuvant biologic therapy is whatever the patient is randomly assigned to in the beginning. They will have patients who will receive a year of lapatinib. In terms of the preoperative portion of the protocols, they’re deliberately similar in design. We’re trying to dovetail the correlative science component so that each trial can serve as a validation of the other.
Track 17
![]() DR LOVE: What do we know about the combination of lapatinib and trastuzumab?
DR LOVE: What do we know about the combination of lapatinib and trastuzumab?
![]() DR CHANG: We have a lot of preclinical data about the synergism
between lapatinib and trastuzumab.
Our group evaluated MCF-7 HER2-overexpressing xenografts and found that with the combination of lapatinib, trastuzumab and endocrine therapy, in ER-positive disease, the tumors all went away. We had almost a 100 percent response rate with the combination of these targeted molecules.
DR CHANG: We have a lot of preclinical data about the synergism
between lapatinib and trastuzumab.
Our group evaluated MCF-7 HER2-overexpressing xenografts and found that with the combination of lapatinib, trastuzumab and endocrine therapy, in ER-positive disease, the tumors all went away. We had almost a 100 percent response rate with the combination of these targeted molecules.
That is strong evidence of cross talk between these different pathways and that pan-HER inhibition is probably necessary.